Single-nucleotide resolution of N6-adenine methylation sites in DNA and RNA by nitrite sequencing†
Abstract
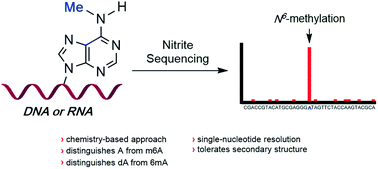
A single-nucleotide resolution sequencing method of N6-adenine methylation sites in DNA and RNA is described. Using sodium nitrite under acidic conditions, chemoselective deamination of unmethylated adenines readily occurs, without competing deamination of N6-adenine sites. The deamination of adenines results in the formation of hypoxanthine bases, which are read by polymerases and reverse transcriptases as guanine; the methylated adenine sites resist deamination and are read as adenine. The approach, when coupled with high-throughput DNA sequencing and mutational analysis, enables the identification of N6-adenine sites in RNA and DNA within various sequence contexts.
- This article is part of the themed collection: 2020 Chemical Science HOT Article Collection


 Please wait while we load your content...
Please wait while we load your content...