Binding of inhibitors to the monomeric and dimeric SARS-CoV-2 Mpro†
Abstract
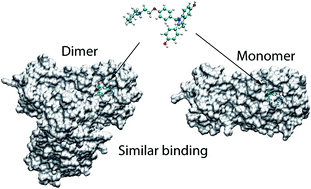
SARS-CoV-2 rapidly infects millions of people worldwide since December 2019. There is still no effective treatment for the virus, resulting in the death of more than one million patients. Inhibiting the activity of SARS-CoV-2 main protease (Mpro), 3C-like protease (3CLP), is able to block the viral replication and proliferation. In this context, our study has revealed that in silico screening for inhibitors of SARS-CoV-2 Mpro can be reliably done using the monomeric structure of the Mpro instead of the dimeric one. Docking and fast pulling of ligand (FPL) simulations for both monomeric and dimeric forms correlate well with the corresponding experimental binding affinity data of 24 compounds. The obtained results were also confirmed via binding pose and noncovalent contact analyses. Our study results show that it is possible to speed up computer-aided drug design for SARS-CoV-2 Mpro by focusing on the monomeric form instead of the larger dimeric one.
- This article is part of the themed collection: Coronavirus articles - free to access collection


 Please wait while we load your content...
Please wait while we load your content...