The structure-based virtual screening of non-benzofuran inhibitors against M. tuberculosis Pks13-TE for anti-tuberculosis phenotypic discovery†
Abstract
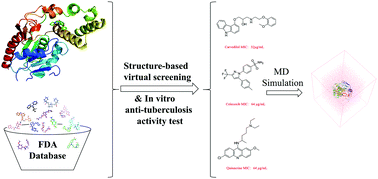
Tuberculosis is the most infectious disease worldwide, primarily because of its increasing resistance to first-line anti-TB drugs. Therefore, development of new drugs with novel mechanisms of action is the solution to the growing problem of drug resistance. The thioesterase domain (TE) of polyketide synthase Pks13 is crucial for the biosynthesis of mycolic acids, which are the essential components of the cell wall of Mycobacterium tuberculosis. However, all the inhibitors of Pks13-TE just share the same benzofuran scaffold, which restricts the further development of Pks13-TE inhibitors. Hence, in order to search for new inhibitors with novel scaffolds and in vitro anti-tuberculosis activity, a structure-based virtual screening method was established to screen the FDA database against pks13-TE. The hit drugs were purchased and tested for anti-tubercular activity against the selectable marker-free autoluminescent Mycobacterium tuberculosis H37Ra (UAlRa). Interestingly, three old drugs approved by FDA, namely, carvedilol (32 μg ml−1), celecoxib (64 μg ml−1), and quinacrine (64 μg ml−1) were discovered to possess in vitro anti-tubercular activity. The complexes of Pks13-TE with the hits showed a similar binding stability and interaction mode with Pks13-TE/co-crystal ligand Tam16, which were validated by the 100 ns molecular dynamics simulations, the binding free energy calculations and alanine scanning mutagenesis analysis. The rest of the chiral isomers for carvedilol and quinacrine were also researched by the same computational strategies. The results of the calculations proved that the S-configuration of carvedilol had a stronger binding affinity with Pks13-TE than the R-configuration of carvedilol, and S and R configurations of quinacrine displayed an equivalent binding affinity with the protein. As the three hit compounds have already been on the market for decades, these compounds possess good enough druggability and definite toxicity and side effects, which were consistent with the prediction results. Our work shows the successful application of computational strategies for the discovery of Pks13-TE non-benzofuran inhibitors with in vitro anti-tubercular activities. These discoveries would further promote the development of Pks13-TE inhibitors and anti-tubercular agents.


 Please wait while we load your content...
Please wait while we load your content...