An optimized culture medium to isolate Lactobacillus fermentum strains from the human intestinal tract†
Abstract
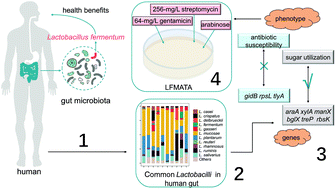
Research studies have shown that Lactobacillus fermentum generally exists in the human gut and has potential health benefits on host health due to its antimicrobial and antioxidant properties. However, the lack of an effective culture medium for the isolation of L. fermentum has presented a significant obstacle on the path to screen L. fermentum strains from the human intestinal tract with a large diversity of commensal microbes. In this study, a total of 51 Lactobacillus species are detected in 200 human fecal samples and we aim to distinguish L. fermentum from these common existing Lactobacillus species and design a more efficient culture medium for isolating L. fermentum strains from the human gut. Based on antibiotic susceptibility and sugar utilization tests, a new optimized medium called LFMATA containing arabinose as the carbon source and 20 mg L−1 vancomycin, 64 mg L−1 gentamicin and 256 mg L−1 streptomycin was developed. Genotype and phenotype analysis for antibiotic resistance and carbohydrate metabolism showed that though glycometabolism-related genes (araA, xylA, manX, bglX, treP and rbsK) correlated with the carbon utilization of Lactobacillus, the genes conferring resistance to streptomycin (gidB and rpsL) and gentamicin (tlyA) were not directly associated with the antibiotic resistance of Lactobacillus strains. This new selective medium greatly increased the efficiency of screening L. fermentum strains from human fecal samples, with the rate of L. fermentum isolation on LFMATA being 10-fold higher than that on LAMVAB.


 Please wait while we load your content...
Please wait while we load your content...