Mass defect-based carbonyl activated tags (mdCATs) for multiplex data-independent acquisition proteome quantification†
Abstract
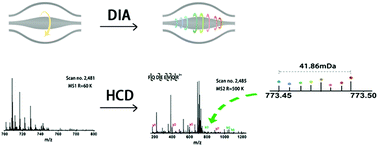
Mass spectrometry (MS) quantification has become an essential tool for both modern biology and translational medicine research. Data-independent acquisition (DIA) has emerged recently and combines the merits of data-dependent acquisition (DDA) methods and selected reaction monitoring (SRM) approaches. Unfortunately, making a quantitative comparison between samples in a single DIA injection is extremely restricted due to its poor compatibility with the traditional DDA labeling approach. As a consequence, a multiplex quantitative reagent suitable for the DIA strategy would be strongly desirable to improve these uninformative situations. As counterparts to traditional DDA quantitative reagents, such as tandem mass tags (TMT) and isobaric tags for relative and absolute quantification (iTRAQ), here, we have designed a novel mass-defect-based carbonyl activated tag (mdCAT) allowing the DIA method to quantify eight samples in parallel for the first time. The integration of the mdCATs with the DIA strategy can not only allow the popular and powerful merits of the DIA strategy, such as broad proteomic coverage, good reproducibility, and accuracy, but it can also eliminate spectral complexity due to their minuscule mass differences. In doing so, it offers new opportunities for the investigation of clinical samples, driving the field of DIA toward unprecedented multiplexing and improving its ability to search for diagnostic biomarkers.


 Please wait while we load your content...
Please wait while we load your content...