Detecting antimicrobial resistance in Escherichia coli using benchtop attenuated total reflectance-Fourier transform infrared spectroscopy and machine learning
Abstract
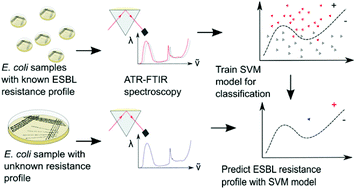
The widespread dissemination of resistance to third-generation cephalosporins in the Enterobacterales through the production of extended-spectrum β-lactamase (ESBL) is considered a critical global crisis requiring urgent attention of clinicians and scientists alike. Rapid diagnostic methods that can identify microbial resistance profiles closer to the point of care are crucial to minimize the overuse of antimicrobial agents and improve patient outcomes. Although Fourier transform infrared (FTIR) microscopy has shown promise in distinguishing between bacterial species, the high cost and technical requirements of the IR microscope may limit broad clinical use. To address the practical needs of a clinical microbiology laboratory, here, we examine the ability of a lower cost portable benchtop attenuated total reflectance (ATR)–FTIR spectrometer to achieve antimicrobial resistance detection, using a simple, clinically aligned sampling protocol. The technical reproducibility was confirmed through multi-day analysis of an Escherichia coli type strain, which serves as quality control. We generated a dataset of 100 E. coli clinical bloodstream isolates with 63 ceftriaxone resistant blaCTX-M ESBL gene variant strains and developed a classifier for blaCTX-M genotype detection. After assessing 35 machine learning methods using the training set (n = 71), four methods were further optimised, and the best performing method was evaluated using the held-out testing set (n = 29). A tuned support vector machine model with a polynomial kernel, using the 700–1500 cm−1 range achieved a sensitivity of 89.2%, and specificity of 66.7% for detecting blaCTX-M in independent testing, approaching the reported performance of FTIR microscopy. With further algorithm improvement, these data suggest the potential deployment of a portable FTIR spectrometer as a rapid antimicrobial susceptibility prediction platform to enable the efficient use of antimicrobials.


 Please wait while we load your content...
Please wait while we load your content...