Deriving accurate molecular indicators of protein synthesis through Raman-based sparse classification†
Abstract
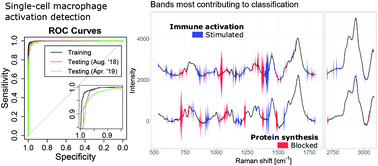
Raman spectroscopy has the ability to retrieve molecular information from live biological samples non-invasively through optical means. Coupled with machine learning, it is possible to use this large amount of information to create models that can predict the state of new samples. We study here linear models, whose separation coefficients can be used to interpret which bands are contributing to the discrimination, and compare the performance of principal component analysis coupled with linear discriminant analysis (PCA/LDA), with regularized logistic regression (Lasso). By applying these methods to single-cell measurements for the detection of macrophage activation, we found that PCA/LDA yields poorer performance in classification compared to Lasso, and underestimates the required sample size to reach stable models. Direct use of Lasso (without PCA) also yields more stable models, and provides sparse separation vectors that directly contain the Raman bands most relevant to classification. To further evaluate these sparse vectors, we apply Lasso to a well-defined case where protein synthesis is inhibited, and show that the separating features are consistent with RNA accumulation and protein levels depletion. Surprisingly, when features are selected purely in terms of their classification power (Lasso), they consist mostly of side bands, while typical strong Raman peaks are not present in the discrimination vector. We propose that this occurs because large Raman bands are representative of a wide variety of intracellular molecules and are therefore less suited for accurate classification.


 Please wait while we load your content...
Please wait while we load your content...