Unfolding mechanism and free energy landscape of single, stable, alpha helices at low pull speeds†
Abstract
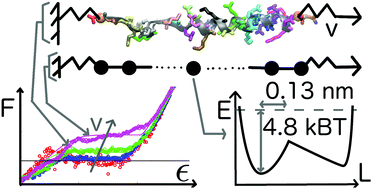
Single alpha helices (SAHs) stable in isolated form are often found in motor proteins where they bridge functional domains. Understanding the mechanical response of SAHs is thus critical to understand their function. The quasi-static force–extension relation of a small number of SAHs is known from single-molecule experiments. Unknown, or still controversial, are the molecular scale details behind those observations. We show that the deformation mechanism of SAHs pulled from the termini at pull speeds approaching the quasi-static limit differs from that of typical helices found in proteins, which are stable only when interacting with other protein domains. Using molecular dynamics simulations with atomistic resolution at low pull speeds previously inaccessible to simulation, we show that SAHs start unfolding from the termini at all pull speeds we investigated. Unfolding proceeds residue-by-residue and hydrogen bond breaking is not the main event determining the barrier to unfolding. We use the molecular simulation data to test the cooperative sticky chain model. This model yields excellent fits of the force–extension curves and quantifies the distance, xE = 0.13 nm, to the transition state, the natural frequency of bond vibration, ν0 = 0.82 ns−1, and the height, V0 = 2.9 kcal mol−1, of the free energy barrier associated with the deformation of single residues. Our results demonstrate that the sticky chain model could advantageously be used to analyze experimental force–extension curves of SAHs and other biopolymers.


 Please wait while we load your content...
Please wait while we load your content...