Target-activated transcription for the amplified sensing of protease biomarkers†
Abstract
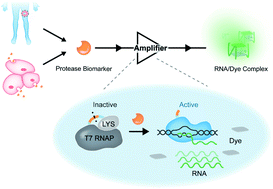
Signal amplification is an effective way to achieve sensitive analysis of biomarkers, exhibiting great promise in biomedical research and clinical diagnosis. Inspired by the transcription process, here we present a versatile strategy that enables effective amplification of proteolysis into nucleic acid signal outputs in a homogeneous system. In this strategy, a protease-activatable T7 RNA polymerase is engineered as the signal amplifier and achieves 3 orders of magnitude amplification in signal gain. The versatility of this strategy has been demonstrated by the development of sensitive and selective assays for protease biomarkers, such as matrix metalloproteinase-2 (MMP-2) and thrombin, with sub-picomole sensitivity, which is 4.3 × 103-fold lower than that of the standard peptide-based method. Moreover, the proposed assay has been further applied in the detection of MMP-2 secreted by cancer cells, as well as in the assessment of MMP-2 levels in osteosarcoma tissue samples, providing a general approach for the monitoring of protease biomarkers in clinical diagnosis.


 Please wait while we load your content...
Please wait while we load your content...