Reversible reconfiguration of high-order DNA nanostructures by employing G-quartet toeholds as adhesive units†
Abstract
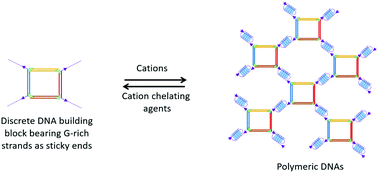
G-quadruplex structures are becoming useful alternative interaction modules for the assembly of DNA nanomaterials because of their unique inducibility by cations. In this study, we demonstrated a new strategy for the assembly of polymeric DNA nanoarchitectures in the presence of cations, such as K+ and Na+, by employing G-quartet toeholds at the edges of discrete mini-square DNA building blocks as adhesive units. In comparison with the Watson–Crick base-paired duplex linkers, G-quadruplex arrays embedded in the self-assembled DNA system exhibit higher thermal stability. The morphology of these doughnut-shaped or spherical-shaped DNA nanostructures is highly regulated by the orientation of the folded G-quadruplexes either in parallel or antiparallel orientation in response to different cations. Furthermore, this G-quadruplex-mediated assembly strategy is able to manipulate the cycling of DNA assemblies between discrete and polymeric states by means of introducing cations and chelating agents sequentially. This property enables the reversible manipulation of the DNA-based nanosystems for at least 4 cycles. The G-quadruplex array embedded in this self-assembled DNA system can become a scaffold for functional molecules, as a number of organic molecules and proteins exhibit specific binding to these G-quadruplex structures. Besides, embedded G-quadruplexes are also considered as functional components of nanoscale electronic materials due to their electron transport through the stacked orientation of the G-quartet. Therefore, this work is an important step towards obtaining reversible, responsive G-quadruplex-induced DNA-based nanomaterials with versatile functionalities which will be highly useful in further electronic, biomedical and drug-delivery applications.


 Please wait while we load your content...
Please wait while we load your content...