A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens†
Abstract
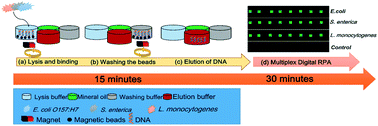
Point-of-care (POC) testing offers rapid diagnostic results. However, the quantification of current methods is performed using standard curves and external references, and not direct and absolute quantification. This paper describes an integrated multiplex digital recombinase polymerase amplification (ImdRPA) microfluidic chip which combines DNA extraction, multiplex digital RPA and fluorescence detection together in one chip, creating a “sample-in-multiplex-digital-answer-out” system. Multi-layer soft lithography technology was used, with polydimethylsiloxane (PDMS) as the chip material and a glass slide as the substrate. This microfluidic chip has a six-layer structure and screw microvalve control function. The sample preparation for the chip involved magnetic bead-based nucleic acid extraction, which was completed within 15 min without any instrument dependence. The dRPA region was divided into 4 regions (3 positive detection areas and 1 negative control area) and included a total of 12 800 chambers, with each chamber being able to contain a volume of 2.7 nL. The screw valve allowed for the reaction components of each specific goal to be pre-embedded in different regions of the chambers. The reagents were passively driven into the dRPA region using vacuum-based self-priming introduction. Furthermore, we successfully demonstrated that the chip can simultaneously detect three species of pathogenic bacteria within 45 min and give digital quantitative results without the need to establish a standard curve in contaminated milk. Moreover, the detection limit of this ImdRPA microfluidic chip was found to be 10 bacterial cells for each kind of pathogen. These characteristics enhance its applicability for rapid detection of foodborne bacteria at the point-of-care (POC). We envision that the further development of this integrated chip will lead to rapid, multiplex and accurate detection of foodborne bacteria in a feasible manner.


 Please wait while we load your content...
Please wait while we load your content...