Microbial genetic potential for xenobiotic metabolism increases with depth during biofiltration†
Abstract
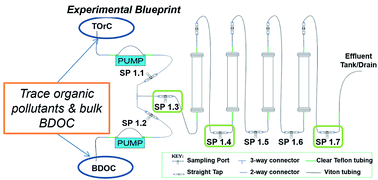
Water infiltration into the subsurface can result in pronounced biogeochemical depth gradients. In this study, we assess metabolic potential and properties of the subsurface microbiome during water infiltration by analyzing sediments from spatially-segmented columns. Past work in these laboratory set-ups demonstrated that removal efficiencies of trace organic pollutants were enhanced by limited availability of biodegradable dissolved organic carbon (BDOC) associated with higher humic ratios and deeper sediment regions. Distinct differences were observed in the microbial community when contrasting shallow versus deeper profile sediments. Metagenomic analyses revealed that shallow sediments contained an enriched potential for bacterial growth and division processes. In contrast, deeper sediments harbored a significant increase in genes associated with the metabolism of secondary metabolites and the biotransformation of xenobiotic water pollutants. Metatranscripts further supported this trend, with increased potential for metabolic attributes associated with the biotransformation of xenobiotics and antibiotic resistance within deeper sediments. Furthermore, increasing ratios of humics in feed solutions correlated to enhanced expression of genes associated with xenobiotic biodegradation. These results provide genetic support for the interplay of dissolved organic carbon limitation and enhanced trace organic biotransformation by the subsurface microbiome.


 Please wait while we load your content...
Please wait while we load your content...