Revealing unfolding steps and volume changes of human telomeric i-motif DNA
Abstract
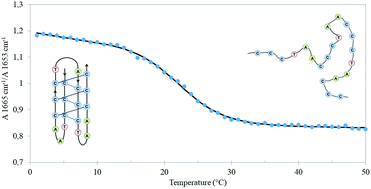
Cytosine rich DNA sequences can fold into a non-canonical four stranded intercalated i-motif structure. We investigated Htel-iM, an i-motif structure of the human telomeric DNA region that plays an important role in cell division, cancer diseases and aging. A high pressure up to 1 Gpa was applied to reveal the volumetric changes during unfolding. Thermal transitions at different pressures were followed by infrared and fluorescence spectroscopy. We demonstrated that Htel-iM unfolds in two steps. First the outer hydrogen bonds break in which C(2)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O(2) and the N(4)H2 amino group is involved, subsequently the hydrogen bond involving N(3)+ becomes broken. Htel-iM was destabilized by pressure and unfolded with a negative volume change.
O(2) and the N(4)H2 amino group is involved, subsequently the hydrogen bond involving N(3)+ becomes broken. Htel-iM was destabilized by pressure and unfolded with a negative volume change.


 Please wait while we load your content...
Please wait while we load your content...