The application of the MM/GBSA method in the binding pose prediction of FGFR inhibitors†
Abstract
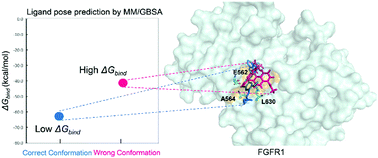
The success of a structure-based drug is highly dependent on a known binding pose of the protein–ligand system. However, this is not always available. In this study, we set out to explore the applicability of the popular and easy-to-use MD-based MM/GBSA method to determine the binding poses of known FGFR inhibitors. It was found that MM/GBSA combined with 100 ns of MD simulation significantly improved the success rate of docking methods from 30–40% to 70%. This work demonstrates a way that the MM/GBSA method can be more accurate than it is in ligand ranking, filling a gap in structure-based drug discovery when the binding pose is unknown.


 Please wait while we load your content...
Please wait while we load your content...