Evaluation of different direct and indirect SELEX monitoring methods and implementation of melt-curve analysis for rapid discrimination of variant aptamer sequences†
Abstract
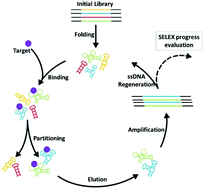
Systematic Evolution of Ligands by Exponential enrichment (SELEX) is an iterative method for in vitro selection of aptamers from a random synthetic oligonucleotide library. Successful retrieving of aptamers by SELEX relies on optimization of various steps including target immobilization, aptamer partitioning, amplification, and ssDNA generation, which all require spending considerable effort and cost. Furthermore, due to the random nature of the initial library, SELEX may redirect toward the selection of low-affinity aptamers that are over-represented in the ssDNA population due to PCR bias. Thus, precise monitoring of the SELEX process is crucial to ensure the selection of target-specific aptamers. In the present study, we investigated the reliability and simplicity of different direct and indirect monitoring methods including UV-Vis spectroscopy, real-time PCR quantification and melt-curve analysis, electrophoretic mobility shift assay (EMSA) and enzyme-linked oligonucleotide assay (ELONA) for selection of DNA aptamers for a protein target. All the examined methods were capable of illustrating the gradual evolution of specific aptamers by the progression of SELEX and showed almost similar results regarding the identification of the enriched round of selection. Moreover, we describe the use of melt-curve analysis in the colony real-time PCR method as a simple, robust, and repeatable tool for pre-sequencing separation of distinct aptamer clones.


 Please wait while we load your content...
Please wait while we load your content...