Mechanical unfolding of alpha- and beta-helical protein motifs†
Abstract
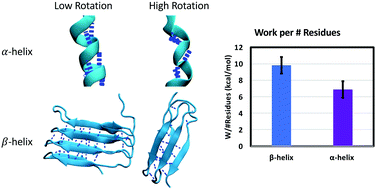
Alpha-helices and beta-sheets are the two most common secondary structure motifs in proteins. Beta-helical structures merge features of the two motifs, containing two or three beta-sheet faces connected by loops or turns in a single protein. Beta-helical structures form the basis of proteins with diverse mechanical functions such as bacterial adhesins, phage cell-puncture devices, antifreeze proteins, and extracellular matrices. Alpha-helices are commonly found in cellular and extracellular matrix components, whereas beta-helices such as curli fibrils are more common as bacterial and biofilm matrix components. It is currently not known whether it may be advantageous to use one helical motif over the other for different structural and mechanical functions. To better understand the mechanical implications of using different helix motifs in networks, here we use Steered Molecular Dynamics (SMD) simulations to mechanically unfold multiple alpha- and beta-helical proteins at constant velocity at the single molecule scale. We focus on the energy dissipated during unfolding as a means of comparison between proteins and work normalized by protein characteristics (initial and final length, # H-bonds, # residues, etc.). We find that although alpha-helices such as keratin and beta-helices CsgA and CsgB can require similar amounts of work to unfold, the normalized work per hydrogen bond, initial end to end length, and number of residues is greater for beta-helices at the same pulling rate. To explain this, we analyze the orientation of the backbone alpha carbons and backbone hydrogen bonds during unfolding. We find that the larger width and shorter height of beta-helices results in smaller angles between the protein backbone and the pulling direction during unfolding. As subsequent strands are separated from the beta-helix core, the angle between the backbone and the pulling direction diminishes. This marks a transition where beta-sheet hydrogen bonds become loaded predominantly in a collective shearing mode, which requires a larger rupture force. This finding underlines the importance of geometry in optimizing resistance to mechanical unfolding in proteins. The helix radius is identified here as an important parameter that governs how much sacrificial energy dissipation capacity can be stored in protein networks, where beta-helices offer unique properties.
- This article is part of the themed collection: Soft Matter Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...