Rational design of a DNA sequence-specific modular protein tag by tuning the alkylation kinetics†
Abstract
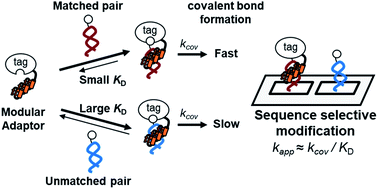
Sequence-selective chemical modification of DNA by synthetic ligands has been a long-standing challenge in the field of chemistry. Even when the ligand consists of a sequence-specific DNA binding domain and reactive group, sequence-selective reactions by these ligands are often accompanied by off-target reactions. A basic principle to design DNA modifiers that react at specific sites exclusively governed by DNA sequence recognition remains to be established. We have previously reported selective DNA modification by a self-ligating protein tag conjugated with a DNA-binding domain, termed as a modular adaptor, and orthogonal application of modular adaptors by relying on the chemoselectivity of the protein tag. The sequence-specific crosslinking reaction by the modular adaptor is thought to proceed in two steps: the first step involves the formation of a DNA–protein complex, while in the second step, a proximity-driven intermolecular crosslinking occurs. According to this scheme, the specific crosslinking reaction of a modular adaptor would be driven by the DNA recognition process only when the dissociation rate of the DNA complex is much higher than the rate constant for the alkylation reaction. In this study, as a proof of principle, a set of combinations for modular adaptors and their substrates were utilized to evaluate the reactions. Three types of modular adaptors consisting of a single type of self-ligating tag and three types of DNA binding proteins fulfill the kinetic requirements for the reaction of the self-ligating tag with a substrate and the dissociation of the DNA–protein complex. These modular adaptors actually undergo sequence-specific crosslinking reactions exclusively driven by the recognition of a specific DNA sequence. The design principle of sequence-specific modular adaptors based on the kinetic aspects of complex formation and chemical modification is applicable for developing recognition-driven selective modifiers for proteins and other biological macromolecules.


 Please wait while we load your content...
Please wait while we load your content...