Single-step multivalent capture assay for nucleic acid detection with dual-affinity regulation using mutation inhibition and allosteric activation†
Abstract
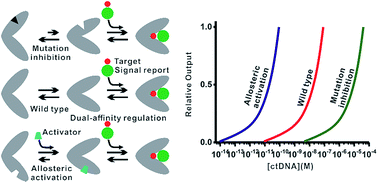
The rational modulation of receptor affinity through distal-site mutation and allosteric control is valuable in biosensor designing to tune the useful dynamic range. Our ability to programmatically engineer dual-affinity regulation into diverse affinities of target binding and activities of hybridization chain reaction, however, remains limited. By programmable engineering of the switching equilibria of the recognition hairpin using distal-site mutation inhibition and allosteric activation, we obtained a set of receptors varying significantly in affinities of target binding and activities of the hybridization chain reaction. For the first time, we developed an electrocatalytic biosensor for nucleic acid detection with a tunable dynamic range based on a conformational switch triggered bidirectional hybridization chain reaction and blocker assisted multivalent binding. This designable biosensor thus enables single-step incubation, diverse affinities of target binding, diverse efficiencies of signal amplification and diverse single nucleotide discrimination for quantitative analyses of nucleic acids of various lengths in serum, which holds great potential as a compelling platform suitable for liquid biopsy.


 Please wait while we load your content...
Please wait while we load your content...