A diffusion-based microfluidic device for single-cell RNA-seq†
Abstract
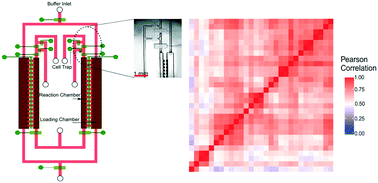
Microfluidic devices provide a low-input and efficient platform for single-cell RNA-seq (scRNA-seq). Existing microfluidic devices have a complicated multi-chambered structure for handling the multi-step process involved in RNA-seq and dilution between steps is used to negate the inhibitory effects among reagents. This makes the device difficult to fabricate and operate. Here we present microfluidic diffusion-based RNA-seq (MID-RNA-seq) for conducting scRNA-seq with a diffusion-based reagent swapping scheme. This device incorporates cell trapping, lysis, reverse transcription and PCR amplification all in one simple microfluidic device. MID-RNA-seq provides high data quality that is comparable to existing scRNA-seq methods while implementing a simple device design that permits multiplexing. The robustness and scalability of the MID-RNA-seq device will be important for transcriptomic studies of scarce cell samples.


 Please wait while we load your content...
Please wait while we load your content...