Resolving complex hierarchies in chemical mixtures: how chemometrics may serve in understanding the immune system
Abstract
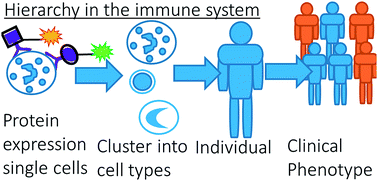
In immunology, the resolution of complex chemical mixtures familiar from omics, comes with an added layer of hierarchy: bioactive immunological surface markers are embedded on the cell membranes of e.g. white blood cells. Therefore, each blood sample actually consists of a comprehensive mixture of cells. The cells need to be resolved based on their surface marker chemistry, to investigate their involvement in an immune response. This mixture may be measured on a single-cell level with Multicolour Flow Cytometry (MFC). Finding such cellular and molecular markers is of the utmost academic and diagnostic importance. Several advanced data analysis methods therefore aim to meet the considerable data challenge of resolving such cell mixtures. These multivariate methods are more resource-efficient than the manual analysis of MFC data, called sequential gating, but also likely provide additional biomedical insight compared to the conventional bivariate approach. To compare such methods more comprehensively than has been done until now, we have developed a list of criteria on how each method recovers the information on both the cell and the underlying molecular levels on an MFC sample of an asthma patient. We compare these methods for the chemometric data analysis commonly used in metabolomics. This shows that all compared methods have their own advantage in recovering the sequential gating results, giving insight into the limitations of sequential gating, providing insight into the chemical relationships between cells within the mixture and resolving information related to chemical heterogeneities between cells. We furthermore show how comparative analyses of different samples may lead to further insight into the subdivision of cells into different types based on their immunological involvement in asthma development, and how sparsity—a currently popular method to enhance the discriminative ability of multivariate models—may reduce the insight into the underlying hierarchical variability in cell chemistry. Although developed for cytometry, the presented chemometrics will be highly valuable to many more chemical systems where hierarchical arrangement of the molecules plays a crucial role.
- This article is part of the themed collection: Challenges in analysis of complex natural mixtures


 Please wait while we load your content...
Please wait while we load your content...