From phage display to structure: an interplay of enthalpy and entropy in the binding of the LDHSLHS polypeptide to silica†
Abstract
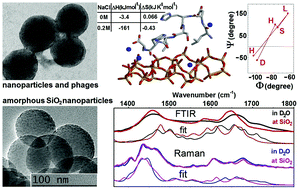
Polypeptide based biosilica composites show promise as next generation multi-functional nano-platforms for diagnostics and bio-catalytic applications. Following the identification of a strong silica binder (LDHSLHS) by phage display, we conduct structural analysis of the polypeptide at the interface with amorphous silica nanoparticles in an aqueous environment. Our approach relies on modelling infrared and Raman spectral responses using predictions of molecular dynamics simulations and quantum studies of the normal modes for several potential structures. By simultaneously fitting both infrared and Raman responses in the amide spectral region, we show that the main structural conformer has a beta-like central region and helix-twisted terminals. Classical simulations, as conducted previously (Chem. Mater., 2014, 26, 5725), predict that the association of the main structure with the interface is stimulated by electrostatic interactions though surface binding also requires spatially distributed sodium ions to compensate for negatively charged acidic silanol groups. Accordingly, diffusion of sodium ions would contribute to a stochastic character of the peptide association with the surface. Consistent with the described dynamics at the interface, the results obtained from isothermal titration calorimetry (ITC) confirm a significant enhancement of polypeptide binding to silica at higher concentrations of Na+. The results of this study suggest that the tertiary structure of a phage capsid protein plays a significant role in regulating the conformation of peptide LDHSLHS, increasing its binding to silica during the phage display process. The results presented here support design-led engineering of polypeptide–silica nanocomposites for bio-technological applications.


 Please wait while we load your content...
Please wait while we load your content...