An SDS-PAGE based proteomic approach for N-terminome profiling†
Abstract
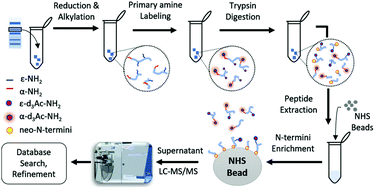
Initial sample quantity, solubilization, separation, and visualization of proteins or their proteolytically altered products are some of the challenges of the currently available solution-based N-termini enrichment methods. We therefore took advantage of the conventional SDS-PAGE system and attempted to address these challenges by proposing a simple yet reproducible, negative selection N-termini enrichment strategy coupled with mass spectrometry based protein identification. It includes in-gel protein level labeling of primary amines using d6-acetic anhydride and post-digestion negative selection of labeled N-terminal peptide(s) using N-hydroxysuccinimide activated agarose beads. We demonstrated the superiority of our method by successfully enriching protein N-termini from as low as 10 ng of bovine serum albumin. The method was validated for its applicability to a complex mixture of proteins by selectively enriching neo-N-termini generated by a site specific protease Glu-C. Its effectiveness for deep N-terminome profiling was also shown using human cell lysate. In addition, a system-wide label-free quantitative proteomic analysis of N-termini in MMP2-perturbed HCT8 cell secretome revealed substrates of several extra- and intra-cellular proteases, which are part of cell growth and proliferation and degradation pathways. In brief, the proposed method demonstrates an effective strategy not only to detect N-termini from a single protein but also for the deep and quantitative analysis of N-terminome from a limited sample amount.

 Please wait while we load your content...
Please wait while we load your content...