Ligand–protein target screening from cell matrices using reactive desorption electrospray ionization-mass spectrometry via a native-denatured exchange approach†
Abstract
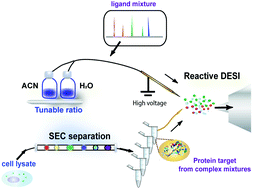
Native mass spectrometry has been recognized as a powerful tool for probing interactions between small molecules, such as drugs and natural products, and target proteins. However, the presence of heterogeneous proteins and metabolites in real biological systems can alter the conformations of target proteins or compete with candidate ligands, thus necessitating a method for measuring binding stoichiometries in matrices aside from the extensively used pure/recombinant protein systems. Furthermore, some small molecule–protein interactions have a transient and low-affinity nature and thus can be mis-assigned as nonspecific binding complexes that are often formed during the native ESI process. A native-denatured exchange (NDX) approach was recently developed using a reactive desorption electrospray ionization-mass spectrometer (DESI-MS) setup to screen specific interacting partners. The method works by gradually increasing the composition of denaturing solvents contained in the DESI spray and thus conferring a switch from a native to denatured ionization environment. This change impairs three-dimensional structures of target proteins and disrupts specific ligand–protein interactions, leading to decreased holo/apo ratios. In contrast, ligand–protein complexes exhibiting different trends are assigned as nonspecific interactions. Herein, we applied the NDX approach to probe specific ligand–protein interactions in biological matrices. We first used mixtures of model ligands and proteins to examine the use of reactive DESI-MS in recognizing ligand–target binding in mixtures. Subsequently, we used the NDX approach to analyze binding affinity curves of ligands to target proteins spiked in cell lysates with the aid of size exclusion chromatography and demonstrated its use in probing specific ligand–protein interactions from cell matrices.


 Please wait while we load your content...
Please wait while we load your content...