Gene specific-loci quantitative and single-base resolution analysis of 5-formylcytosine by compound-mediated polymerase chain reaction†
Abstract
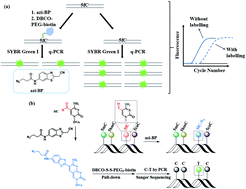
5-Formylcytosine (5fC) is known as one of the key players in the process of active DNA demethylation and displays essential epigenetic functions in mammals. In spite of the blooming development of whole genome sequencing methods for this modified cytosine base, the easily operated gene specific-loci detection of 5fC has rarely been reported. Herein, we present a compound-mediated analysis of the content and site of 5fC by the polymerase chain reaction (PCR) assay. The molecule, namely azi-BP, which can selectively label 5fC and form a huge group through a click chemistry reaction, hindering the amplification activity of Taq DNA polymerase, acts as a “roadblock” and enables the quantitative analysis of 5fC by quantitative polymerase chain reaction (qPCR). The existence of 5fC in several fragment-specific genomic DNAs of mouse embryonic stem cells (mESCs) was successfully demonstrated using this method. In addition, the gene fragment containing 5fC can be easily biotinylated and enriched after labeling with azi-BP. Moreover, after azi-BP incorporation, the loss of the exocyclic 4-amino group of 5fC leads to C-to-T conversion and subsequent pairing with adenine (A) in the PCR, which can accurately identify 5fC sites at single-base resolution by site-specific mutation. Azi-BP shows high selectivity to 5fC among all modified pyrimidine bases, revealing that this compound-mediated assay can be applied for content and single-base resolution analysis for gene specific-loci of 5fC.


 Please wait while we load your content...
Please wait while we load your content...