Identification of pathogenic bacteria in complex samples using a smartphone based fluorescence microscope†
Abstract
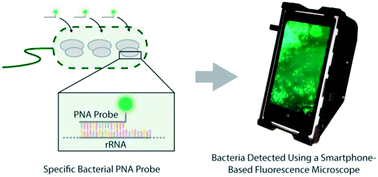
Diagnostics based on fluorescence imaging of biomolecules is typically performed in well-equipped laboratories and is in general not suitable for remote and resource limited settings. Here we demonstrate the development of a compact, lightweight and cost-effective smartphone-based fluorescence microscope, capable of detecting signals from fluorescently labeled bacteria. By optimizing a peptide nucleic acid (PNA) based fluorescence in situ hybridization (FISH) assay, we demonstrate the use of the smartphone-based microscope for rapid identification of pathogenic bacteria. We evaluated the use of both a general nucleic acid stain as well as species-specific PNA probes and demonstrated that the mobile platform can detect bacteria with a sensitivity comparable to that of a conventional fluorescence microscope. The PNA-based FISH assay, in combination with the smartphone-based fluorescence microscope, allowed us to qualitatively analyze pathogenic bacteria in contaminated powdered infant formula (PIF) at initial concentrations prior to cultivation as low as 10 CFU per 30 g of PIF. Importantly, the detection can be done directly on the smartphone screen, without the need for additional image analysis. The assay should be straightforward to adapt for bacterial identification also in clinical samples. The cost-effectiveness, field-portability and simplicity of this platform will create various opportunities for its use in resource limited settings and point-of-care offices, opening up a myriad of additional applications based on other fluorescence-based diagnostic assays.
- This article is part of the themed collection: Editors’ collection: Fluorescent Sensors


 Please wait while we load your content...
Please wait while we load your content...