SD-chip enabled quantitative detection of HIV RNA using digital nucleic acid sequence-based amplification (dNASBA)†
Abstract
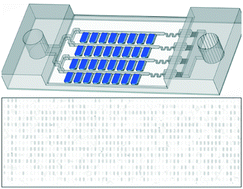
Quantitative detection of RNA is important in molecular biology and clinical diagnostics. Nucleic acid sequence-based amplification (NASBA), a single-step method to amplify single-stranded RNA, is attractive for use in point-of-care (POC) diagnostics because it is an isothermal technique that is as sensitive as RT-PCR with a shorter reaction time. However, NASBA is limited in its ability to provide accurate quantitative information, such as viral load or RNA copy number. Here we test a digital format of NASBA (dNASBA) using a self-digitization (SD) chip platform, and apply it to quantifying HIV-1 RNA. We demonstrate that dNASBA is more sensitive and accurate than the real-time quantitative NASBA, and can be used to quantify HIV-1 RNA in plasma samples. Digital NASBA is thus a promising POC diagnostics tool for use in resource-limited settings.


 Please wait while we load your content...
Please wait while we load your content...