Molecular dynamics modeling of Pseudomonas aeruginosa outer membranes†
Abstract
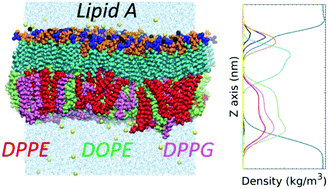
Pseudomonas aeruginosa is a common Gram-negative bacterium and opportunistic human pathogen. The distinctive structure of its outer membrane (OM) and outer membrane vesicles (OMVs) plays a fundamental role in bacterial virulence, colonization ability, and antibiotic resistance. To provide critical insights into OM and OMV functionality, we conducted an all-atom molecular dynamics study of asymmetric membranes that are biologically relevant to P. aeruginosa. We hybridized a GLYCAM06-based lipopolysaccharides force field with the Stockholm lipids force field (Slipids) to model bilayer membranes with Lipid A molecules in one leaflet and physiologically relevant phospholipid molecules in the other, including 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), and 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG). In particular, a membrane with phospholipid composition representing the P. aeruginosa OM was constructed and modeled by mixing the physiologically dominant components. The detailed structure of membranes was characterized by area per lipid, transmembrane mass and charge densities, radial distribution function (RDF), deuterium order parameter (SCD) of acyl chains, and inclination angles of phosphates and disaccharide in Lipid A. The membrane fluidity in equilibrium and the hydration of functional groups were probed and characterized quantitatively. The consistent properties of the Lipid A leaflets in different membranes demonstrate its compatibility with various phospholipids present in the P. aeruginosa OM. The more ordered acyl chains of Lipid A compared to the cytoplasmic cell membrane contribute to the low permeability of bacterial outer membrane. The findings of this computational investigation of P. aeruginosa OM will further the understanding of microbial pathogenesis and enable future study of OMV biogenesis.


 Please wait while we load your content...
Please wait while we load your content...