Protein environment affects the water–tryptophan binding mode. MD, QM/MM, and NMR studies of engrailed homeodomain mutants†
Abstract
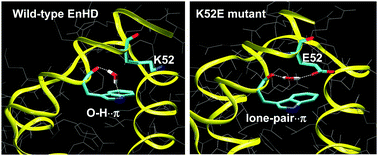
Water molecules can interact with aromatic moieties using either their O–H bonds or their lone-pairs of electrons. In proteins, water–π interactions have been reported to occur with tryptophan and histidine residues, and dynamic exchange between O–H⋯π hydrogen bonding and lone-pair⋯π interactions was suggested to take place, based on ab initio calculations. Here we used classical and QM/MM molecular dynamics simulations, complemented with an NMR study, to examine a specific water–indole interaction observed in the engrailed homeodomain and in its mutants. Our simulations indicate that the binding mode between water and indole can adapt to the potential created by the surrounding amino acids (and by the residues at the DNA surface in protein–DNA complexes), and support the model of dynamic switching between the O–H⋯π hydrogen bonding and lone-pair⋯π binding modes.


 Please wait while we load your content...
Please wait while we load your content...