A G-quadruplex motif at the 3′ end of sgRNAs improves CRISPR–Cas9 based genome editing efficiency†
Abstract
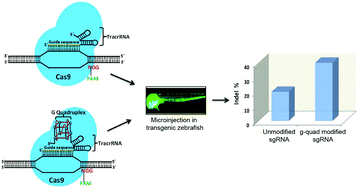
Originating as a component of prokaryotic adaptive immunity, the type II CRISPR/Cas9 system has been repurposed for targeted genome editing in various organisms. Although Cas9 can bind and cleave DNA efficiently under in vitro conditions, its activity inside a cell can vary dramatically between targets owing to the differences between genomic loci and the availability of enough Cas9/sgRNA (single guide RNA) complex molecules for cleavage. Most methods have so far relied on Cas9 protein engineering or base modifications in the sgRNA sequence to improve CRISPR/Cas9 activity. Here we demonstrate that a structure based rational design of sgRNAs can enhance the efficiency of Cas9 cleavage in vivo. By appending a naturally forming RNA G-quadruplex motif to the 3′ end of sgRNAs we can improve its stability and target cleavage efficiency in zebrafish embryos without inducing off-target activity, thereby underscoring its value in the design of better and optimized genome editing triggers.


 Please wait while we load your content...
Please wait while we load your content...