RNA splicing process analysis for identifying antisense oligonucleotide inhibitors with padlock probe-based isothermal amplification†
Abstract
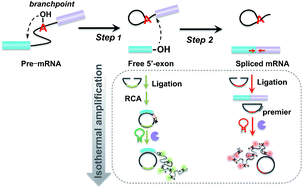
RNA splicing, which mainly involves two transesterification steps, is a fundamental process of gene expression and its abnormal regulation contributes to serious genetic diseases. Antisense oligonucleotides (ASOs) are genetic control tools that can be used to specifically control genes through alteration of the RNA splicing pathway. Despite intensive research, how ASOs or various other factors influence the multiple processes of RNA splicing still remains obscure. This is largely due to an inability to analyze the splicing efficiency of each step in the RNA splicing process with high sensitivity. We addressed this limitation by introducing a padlock probe-based isothermal amplification assay to achieve quantification of the specific products in different splicing steps. With this amplified assay, the roles that ASOs play in RNA splicing inhibition in the first and second steps could be distinguished. We identified that 5′-ASO could block RNA splicing by inhibiting the first step, while 3′-ASO could block RNA splicing by inhibiting the second step. This method provides a versatile tool for assisting efficient ASO design and discovering new splicing modulators and therapeutic drugs.


 Please wait while we load your content...
Please wait while we load your content...