Real-time activity assays of β-lactamases in living bacterial cells: application to the inhibition of antibiotic-resistant E. coli strains†
Abstract
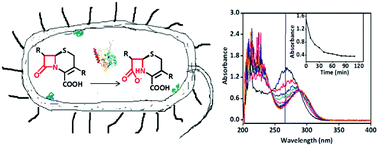
The emergence of antibiotic resistance caused by β-lactamases, including serine β-lactamases (SβLs) and metallo-β-lactamases (MβLs), is a global public health threat. L1, a B3 subclass MβL, hydrolyzes almost all of known β-lactam antibiotics. We report a simple and straightforward UV-Vis approach for real-time activity assays of β-lactamases inside living bacterial cells, and this method has been exemplified by choosing antibiotics, L1 enzyme, Escherichia coli expressing L1 (L1 E. coli), Escherichia coli expressing extended-spectrum β-lactamases (ESBL-E. coli), clinical bacterial strains, and reported MβL and SβL inhibitors. The cell-based studies demonstrated that cefazolin was hydrolyzed by L1 E. coli and clinical strains, and confirmed the hydrolysis to be inhibited by two known L1 inhibitors EDTA and azolylthioacetamide (ATAA), with an IC50 value of 1.6 and 18.9 μM, respectively. Also, it has been confirmed that the breakdown of cefazolin caused by ESBL-E. coli was inhibited by clavulanic acid, the first SβL inhibitor approved by FDA. The data gained through this approach are closely related to the biological function of the target enzyme in its physiological environment. The UV-Vis method proposed here can be applied to target-based whole-cell screening to search for potent β-lactamase inhibitors, and to assays of reactions in complex biological systems, for instance in medical assays.


 Please wait while we load your content...
Please wait while we load your content...