iMulti-HumPhos: a multi-label classifier for identifying human phosphorylated proteins using multiple kernel learning based support vector machines†
Abstract
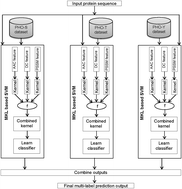
Protein phosphorylation plays a potential role in regulating protein conformation and functions. As a result, identifying an uncharacterized protein sequence as a phosphorylated protein is a very meaningful problem and an urgent issue for both basic research and drug development. Although various types of computational methods have been developed to identify the phosphorylation sites for a recognized phosphorylated protein, very few computational methods have been developed to identify whether an uncharacterized protein can be phosphorylated or not. Therefore, there exists some scope for further improvement to characterize a protein as phosphorylated or not. Among all the residues of protein molecules, three types of amino acid residues, namely serine, threonine, and tyrosine, have been found to be susceptible to phosphorylation, which leads to the requirement of multi-label phosphorylated protein identification. Therefore, in this study, a novel computational tool termed iMulti-HumPhos has been developed to predict multi-label phosphorylated proteins by (1) extracting three different sets of features from protein sequences, (2) defining an individual kernel for each set of features and combining them into a single kernel using multiple kernel learning, and (3) constructing a multi-label predictor using a combination of support vector machines (SVMs) where each SVM has been trained with the combined kernel. In addition, we have balanced the effect of the skewed training dataset by the Different Error Costs method for the development of our system. The experimental results show that the iMulti-HumPhos predictor provides significantly better performance than the existing predictor Multi-iPPseEvo. A user-friendly web-server of iMulti-HumPhos is available at http://research.ru.ac.bd/iMulti-HumPhos/.


 Please wait while we load your content...
Please wait while we load your content...