Structure of the H-NS–DNA nucleoprotein complex†
Abstract
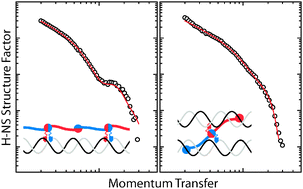
Nucleoid associated proteins (NAPs) play a key role in the compaction and expression of the prokaryotic genome. Here we report the organisation of a major NAP, the protein H-NS on a double stranded DNA fragment. For this purpose we have carried out a small angle neutron scattering study in conjunction with contrast variation to obtain the contributions to the scattering (structure factors) from DNA and H-NS. The H-NS structure factor agrees with a heterogeneous, two-state binding model with sections of the DNA duplex surrounded by protein and other sections having protein bound to the major groove. In the presence of magnesium chloride, we observed a structural rearrangement through a decrease in cross-sectional diameter of the nucleoprotein complex and an increase in fraction of major groove bound H-NS. The two observed binding modes and their modulation by magnesium ions provide a structural basis for H-NS-mediated genome organisation and expression regulation.

 Please wait while we load your content...
Please wait while we load your content...