Disruption of protein–protein interactions: hot spot detection, structure-based virtual screening and in vitro testing for the anti-cancer drug target – survivin†
Abstract
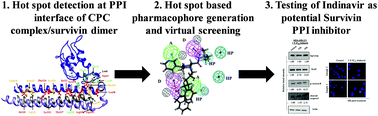
Survivin is a member of the inhibitor of the apoptosis (IAP) family of proteins, and plays a crucial role in both cell division and apoptosis. As it is overexpressed in many human solid tumors, it has become an attractive drug target for cancer therapy. Survivin is involved in protein–protein interactions (PPI) with several of its substrate proteins, and disruption of these interactions could be a possible means to target the function of survivin in cancer. To this end, we sought and were able to detect hot spot residues in the survivin dimer and Chromosomal Passenger Complex (CPC; survivin/borealin/INCENP) using simple knowledge based physical models implemented in the Robetta server (http://robetta.bakerlab.org/), KFC server (http://kfc.mitchell-lab.org/) and the HotRegion database (http://prism.ccbb.ku.edu.tr/hotregion/). Then, extensive molecular dynamics simulations were applied to generate an ensemble of conformations and were used to quantitatively estimate the binding free energy of the identified hot spot residues using MM-PBSA alanine scanning mutagenesis and per-residue energy decomposition. Based on the frequency of occurrence of the hot spot residues and the estimated binding free energy, the survivin dimer and CPC interface residues were designated as “hot spots” and “warm spots”. Finally, based on the identified hot spots of survivin (Leu6A, Trp10A, Leu98A, Phe101A, Asp105A, and Arg106A), a pharmacophore model was derived and used to virtually screen database compounds to identify indinavir as potential inhibitor that could target survivin PPI. A preliminary biochemical investigation shows that the treatment of MDA-MB-231 breast cancer cells with indinavir resulted in Aurora B and XIAP downregulation and caspase-3 activation, hallmarks of survivin PPI inhibition.

 Please wait while we load your content...
Please wait while we load your content...