Nucleobase-functionalized graphene nanoribbons for accurate high-speed DNA sequencing†
Abstract
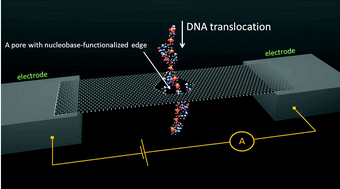
We propose a water-immersed nucleobase-functionalized suspended graphene nanoribbon as an intrinsically selective device for nucleotide detection. The proposed sensing method combines Watson–Crick selective base pairing with graphene's capacity for converting anisotropic lattice strain to changes in an electrical current at the nanoscale. Using detailed atomistic molecular dynamics (MD) simulations, we study sensor operation at ambient conditions. We combine simulated data with theoretical arguments to estimate the levels of measurable electrical signal variation in response to strains and determine that the proposed sensing mechanism shows significant promise for realistic DNA sensing devices without the need for advanced data processing, or highly restrictive operational conditions.

 Please wait while we load your content...
Please wait while we load your content...