Thermal unfolding simulations of NBD1 domain variants reveal structural motifs associated with the impaired folding of F508del-CFTR†
Abstract
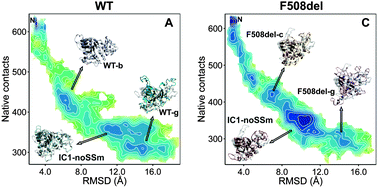
We employed high-temperature classical molecular dynamics (MD) simulations to investigate the unfolding process of the wild-type (WT) and F508del-NBD1 domains of CFTR protein, with and without second-site mutations. To rationalize the in vitro behavior of F508del-NBD1, namely its lower folding yield and higher aggregation propensity, we focused our analysis of the MD data on the existence of intermediate states with aggregation potential and/or stabilized by a significant number of non-native interactions (i.e. misfolded states). We find that the deletion of phenylalanine 508 is able to forcefully reshape the conformational space of the NBD1 domain to the extent that it uniquely populates intermediate states whose structural traits provide important insights into the molecular events that underlie the impaired folding of F508del-NBD1. In particular, our simulations predict the formation of a misfolded intermediate whose population is highly enhanced by deletion of residue 508. The stabilization of this intermediate is a direct consequence of the enhanced non-native coupling between various key regions of the α-helical subdomain and ATP-binding subdomain; it is singularly characterized by a set of non-native interactions within the ATP-binding subdomain and between that domain and the α-helical subdomain region. The formation of this intermediate is not blocked by second-site suppressor mutations which indicates a limited role of the latter in correcting the rather complex folding process of the CFTR protein missing residue 508.

 Please wait while we load your content...
Please wait while we load your content...