Dynamics of GCN4 facilitate DNA interaction: a model-free analysis of an intrinsically disordered region†
Abstract
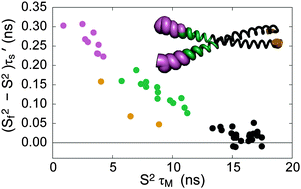
Intrinsically disordered proteins (IDPs) and proteins with intrinsically disordered regions (IDRs) are known to play important roles in regulatory and signaling pathways. A critical aspect of these functions is the ability of IDP/IDRs to form highly specific complexes with target molecules. However, elucidation of the contributions of conformational dynamics to function has been limited by challenges associated with structural heterogeneity of IDP/IDRs. Using NMR spin relaxation parameters (15N R1, 15N R2, and {1H}–15N heteronuclear NOE) collected at four static magnetic fields ranging from 14.1 to 21.1 T, we have analyzed the backbone dynamics of the basic leucine-zipper (bZip) domain of the Saccharomyces cerevisiae transcription factor GCN4, whose DNA binding domain is intrinsically disordered in the absence of DNA substrate. We demonstrate that the extended model-free analysis can be applied to proteins with IDRs such as apo GCN4 and that these results significantly extend previous NMR studies of GCN4 dynamics performed using a single static magnetic field of 11.74 T [Bracken, et al., J. Mol. Biol., 1999, 285, 2133–2146] and correlate well with molecular dynamics simulations [Robustelli, et al., J. Chem. Theory Comput., 2013, 9, 5190–5200]. In contrast to the earlier work, data at multiple static fields allows the time scales of internal dynamics of GCN4 to be reliably quantified. Large amplitude dynamic fluctuations in the DNA-binding region have correlation times (τs ≈ 1.4–2.5 ns) consistent with a two-step mechanism in which partially ordered bZip conformations of GCN4 form initial encounter complexes with DNA and then rapidly rearrange to the high affinity state with fully formed basic region recognition helices.
- This article is part of the themed collection: Exploring the conformational heterogeneity of biomolecules: theory and experiments

 Please wait while we load your content...
Please wait while we load your content...