Connecting localized DNA strand displacement reactions†
Abstract
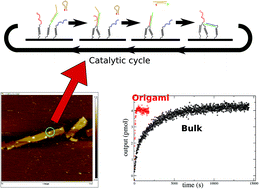
Logic circuits based on DNA strand displacement reactions have been shown to be versatile enough to compute the square root of four-bit numbers. The implementation of these circuits as a set of bulk reactions faces difficulties which include leaky reactions and intrinsically slow, diffusion-limited reaction rates. In this paper, we consider simple examples of these circuits when they are attached to platforms (DNA origamis). As expected, constraining distances between DNA strands leads to faster reaction rates. However, it also induces side-effects that are not detectable in the solution-phase version of this circuitry. Appropriate design of the system, including protection and asymmetry between input and fuel strands, leads to a reproducible behaviour, at least one order of magnitude faster than the one observed under bulk conditions.

 Please wait while we load your content...
Please wait while we load your content...