A novel method for predicting post-translational modifications on serine and threonine sites by using site-modification network profiles†
Abstract
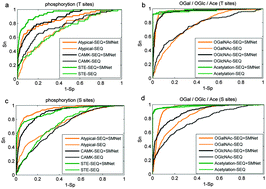
Post-translational modifications (PTMs) regulate many aspects of biological behaviours including protein–protein interactions and cellular processes. Identification of PTM sites is helpful for understanding the PTM regulatory mechanisms. The PTMs on serine and threonine sites include phosphorylation, O-linked glycosylation and acetylation. Although a lot of computational approaches have been developed for PTM site prediction, currently most of them generate the predictive models by employing only local sequence information and few of them consider the relationship between different PTMs. In this paper, by adopting the site-modification network (SMNet) profiles that efficiently incorporate in situ PTM information, we develop a novel method to predict PTM sites on serine and threonine. PTM data are collected from various PTM databases and the SMNet is built to reflect the relationship between multiple PTMs, from which SMNet profiles are extracted to train predictive models based on SVM. Performance analysis of the SVM models shows that the SMNet profiles play an important role in accurately predicting PTM sites on serine and threonine. Furthermore, the proposed method is compared with existing PTM prediction approaches. The results from 10-fold cross-validation demonstrate that the proposed method with SMNet profiles performs remarkably better than existing methods, suggesting the power of SMNet profiles in identifying PTM sites.

 Please wait while we load your content...
Please wait while we load your content...