An immunomics approach for the analysis of natural antibody responses to Plasmodium vivax infection†
Abstract
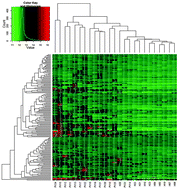
High throughput immunomics is a powerful platform to discover potential targets of host immunity and develop diagnostic tests for infectious diseases. We screened the sera of Plasmodium vivax-exposed individuals to profile the antibody response to blood-stage antigens of P. vivax using a P. vivax protein microarray. A total of 1936 genes encoding the P. vivax proteins were expressed, printed and screened with sera from P. vivax-exposed individuals and normal subjects. Total of 151 (7.8% of the 1936 targets) highly immunoreactive antigens were identified, including five well-characterized antigens of P. vivax (ETRAMP11.2, Pv34, SUB1, RAP2 and MSP4). Among the highly immunoreactive antigens, 5 antigens were predicted as adhesins by MAAP, and 11 antigens were predicted as merozoite invasion-related proteins based on homology with P. falciparum proteins. There are 40 proteins that have serodiagnostic potential for antibody surveillance. These novel Plasmodium antigens identified provide the clues for understanding host immune response to P. vivax infection and the development of antibody surveillance tools.

 Please wait while we load your content...
Please wait while we load your content...