Pseudo nucleotide composition or PseKNC: an effective formulation for analyzing genomic sequences
Abstract
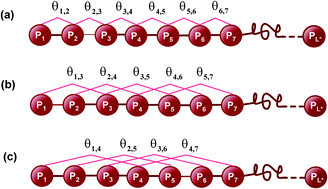
With the avalanche of DNA/RNA sequences generated in the post-genomic age, it is urgent to develop automated methods for analyzing the relationship between the sequences and their functions. Towards this goal, a series of sequence-based methods have been proposed and applied to analyze various character-unknown DNA/RNA sequences in order for in-depth understanding their action mechanisms and processes. Compared with the classical sequence-based methods, the pseudo nucleotide composition or PseKNC approach developed very recently has the following advantages: (1) it can convert length-different DNA/RNA sequences into dimension-fixed digital vectors that can be directly handled by all the existing machine-learning algorithms or operation engines; (2) it can contain the desired features and properties according to the selection or definition of users; (3) it can cover considerable sequence pattern information, both local and global. This minireview is focused on the concept of pseudo nucleotide composition, its development and applications.
- This article is part of the themed collection: 10th Anniversary of Molecular BioSystems

 Please wait while we load your content...
Please wait while we load your content...