Predicting the binding modes and sites of metabolism of xenobiotics†
Abstract
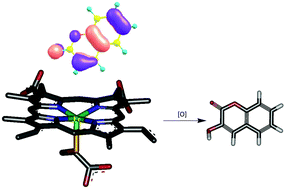
Metabolism studies are an essential integral part of ADMET profiling of drug candidates to evaluate their safety and efficacy. Cytochrome P-450 (CYP) metabolizes a wide variety of xenobiotics/drugs. The binding modes of these compounds with CYP and their intrinsic reactivities decide the metabolic products. We report here a novel computational protocol, which comprises docking of ligands to heme-containing CYPs and prediction of binding energies through a newly developed scoring function, followed by analyses of the docked structures and molecular orbitals of the ligand molecules, for predicting the sites of metabolism (SOM) of ligands. The calculated binding free energies of 121 heme-containing protein–ligand docked complexes yielded a correlation coefficient of 0.84 against experiment. Molecular orbital analyses of the resultant top three unique poses of the docked complexes provided a success rate of 87% in identifying the experimentally known sites of metabolism of the xenobiotics. The SOM prediction methodology is freely accessible at http://www.scfbio-iitd.res.in/software/drugdesign/som.jsp.

 Please wait while we load your content...
Please wait while we load your content...