Construction and analysis of a protein–protein interaction network related to self-renewal of mouse spermatogonial stem cells†
Abstract
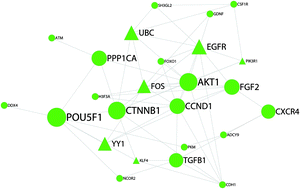
Spermatogonial stem cells (SSCs) are responsible for sustained spermatogenesis throughout the reproductive life of the male. Extensive studies of SSCs have identified dozens of genes that play important roles in sustaining or controlling the pool of SSCs in the mammalian testis. However, there is still limited knowledge of whether or how these key genes interact with each other during SSC self-renewal. Here, we constructed a protein–protein interaction (PPI) network for SSC self-renewal based on interactions between 23 genes essential for SSC self-renewal, which were obtained from a text mining system, and the interacting partners of the 23 key genes, which were differentially expressed in SSCs. The SSC self-renewal PPI network consisted of 246 nodes connected by 844 edges. Topological analyses of the PPI network were conducted to identify genes essential for maintenance of SSC self-renewal. The subnetwork of the SSC self-renewal network suggested that the 23 key genes involved in SSC self-renewal were connected together through other 94 genes. Clustering of the whole network and subnetwork of SSC self-renewal revealed several densely connected regions, implying significant molecular interaction modules essential for SSC self-renewal. Notably, we found the 23 genes to be responsible for SSC self-renewal by forming a continuous PPI network centered on Pou5f1. Our study indicates that it is feasible to explore important proteins and regulatory pathways in biological activities by combining a PPI database with the high-throughput data of gene expression profiles.

 Please wait while we load your content...
Please wait while we load your content...