Metabolic faecal fingerprinting of trans-resveratrol and quercetin following a high-fat sucrose dietary model using liquid chromatography coupled to high-resolution mass spectrometry†
Abstract
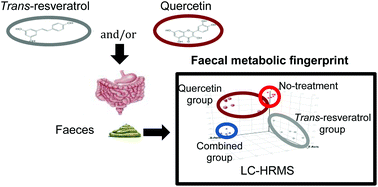
Faecal non-targeted metabolomics deciphers metabolic end-products resulting from the interactions among food, host genetics, and gut microbiota. Faeces from Wistar rats fed a high-fat sucrose (HFS) diet supplemented with trans-resveratrol and quercetin (separately or combined) were analysed by liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS). Metabolomics in faeces are categorised into four clusters based on the type of treatment. Tentative identification of significantly differing metabolites highlighted the presence of carbohydrate derivatives or conjugates (3-phenylpropyl glucosinolate and dTDP-D-mycaminose) in the quercetin group. The trans-resveratrol group was differentiated by compounds related to nucleotides (uridine monophosphate and 2,4-dioxotetrahydropyrimidine D-ribonucleotide). Marked associations between bacterial species (Clostridium genus) and the amount of some metabolites were identified. Moreover, trans-resveratrol and resveratrol-derived microbial metabolites (dihydroresveratrol and lunularin) were also identified. Accordingly, this study confirms the usefulness of omics-based techniques to discriminate individuals depending on the physiological effect of food constituents and represents an interesting tool to assess the impact of future personalized therapies.

 Please wait while we load your content...
Please wait while we load your content...