Impacts of retinal polyene (de)methylation on the photoisomerization mechanism and photon energy storage of rhodopsin†
Abstract
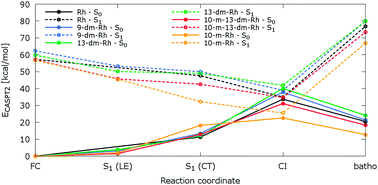
Ab initio multiconfigurational quantum chemical methodology combined with molecular mechanics (CASPT2//CASSCF/AMBER) was applied to probe impacts of retinal protonated Schiff base (RPSB) polyene methylation and/or demethylation on the mechanism of photochemical isomerization in bovine rhodopsin. We have examined structural and spectroscopic properties of wild-type rhodopsin (with 11-cis-9,13-dimethyl-RPSB) and artificial rhodopsins, hosting four 11-cis-RPSB derivatives, 13-demethyl-, 9-demethyl-, 10-methyl-13-demethyl-, and 10-methyl-RPSB, evolving along the photoisomerization coordinate. It is found that the addition of 10-methyl or/and deletion of 9-/13-methyl groups do not appear to interfere structurally with the photoisomerization pathway in the S1 excited state. Remarkably, the two-mode space-saving mechanism initiated by bond order inversion and followed by asynchronous bicycle-pedal distortion in the RPSB backbone drives the photoreaction in all rhodopsin analogues studied here. However, methylation and/or demethylation is responsible for perturbation of excess energy deposited in the conical intersection structures. The analysis of photon energy stored by bathorhodopsin in synthetic pigments reveals that it is affected by steric crowding of methyl substituents in the RPSB backbone.

 Please wait while we load your content...
Please wait while we load your content...