Dimer domain swapping versus monomer folding in apo-myoglobin studied by molecular simulations
Abstract
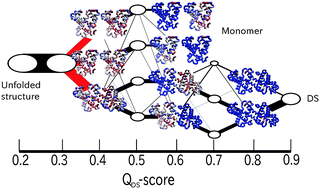
Recent experiments showed that myoglobin can form a domain-swapped dimer under certain conditions. Here, using a coarse-grained symmetrized Go model, we performed a series of folding simulations of two apo-myoglobin molecules restrained at a high density, addressing competition of formation of a domain-swapped dimer with folding to two monomer structures. In addition to the domain-swapped dimer found in X-ray crystallography, we also found some other forms of domain swapping. With the protein density, the probability of domain-swapping increased. Folding pathway analysis clarified that separation between monomer folding and domain-swapping arose at a relatively early stage, where inter-chain contacts between helices AB of one chain and helices GH of another chain tend to result in the domain swapped dimer. This resembles the mechanism of domain swapping suggested previously for cytochrome c.

 Please wait while we load your content...
Please wait while we load your content...