Graphene nanoflakes as an efficient ionizing matrix for MALDI-MS based lipidomics of cancer cells and cancer stem cells†
Abstract
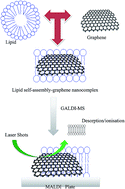
This study demonstrates that graphene nanoflakes can be efficiently used as a successful, interference free matrix for matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). The method is referred to as graphene-assisted laser desorption/ionization mass spectrometry (GALDI-MS), and it can be used for lipidomic analysis of cancer cells and cancer stem cells. The graphene nanoflakes were synthesized in-house and exhibited a transparent flake-like shape with characteristic crumpled silk waves in transmission electron microscopy and typical absorption characteristics upon UV-vis (λmax = 270 nm) and Fourier-transform infrared spectroscopic analysis. Graphene nanoflakes were tested as a sole matrix and co-matrix with traditional MALDI-MS matrices for the lipid extracts obtained from normal breast, cancer and cancer stem cells with four different spotting methods. In all the cases, the graphene nanoflakes displayed a noise free and good quality mass spectrum. This study reveals the possibility that lipids could self-assemble as a multi-layered structure on the graphene nanoflake platform by electrostatic interactions between the graphene nanoflakes and the lipid head groups and thus result in noise-free spectra from GALDI-MS based lipidomics.

 Please wait while we load your content...
Please wait while we load your content...