Illuminating HIV gp120-ligand recognition through computationally-driven optimization of antibody-recruiting molecules†
Abstract
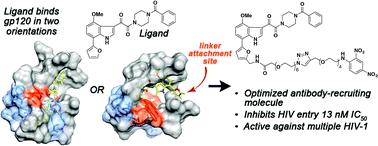
Here we report on the structure-based optimization of antibody-recruiting molecules targeting HIV gp120 (ARM-H). These studies have leveraged a combination of medicinal chemistry, biochemical and cellular assay analysis, and computation. Our findings have afforded an optimized analog of ARM-H, which is ∼1000 fold more potent in gp120-binding and MT-2 antiviral assays than our previously reported derivative. Furthermore, computational analysis, taken together with experimental data, provides evidence that azaindole- and indole-based attachment inhibitors bind gp120 at an accessory hydrophobic pocket beneath the CD4-binding site and can also adopt multiple distinct binding modes in interacting with gp120. These results are likely to prove enabling in the development of novel HIV attachment inhibitors, and more broadly, they suggest novel applications for ARMs as probes of conformationally flexible systems.

 Please wait while we load your content...
Please wait while we load your content...