Paper microfluidic extraction and direct smartphone-based identification of pathogenic nucleic acids from field and clinical samples
Abstract
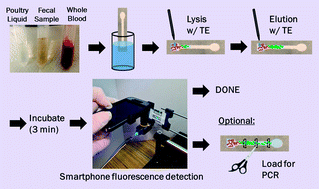
A rapid, paper microfluidic- and smartphone-based protocol was developed for the extraction and direct fluorescent identification of the nucleic acids of Salmonella Typhimurium from field and clinical samples. Initially, liquid samples (10% diluted) from fresh poultry packaging were loaded on the paper chips and were lysed with Tris–EDTA (TE) buffer. Nucleic acids from the lysed samples were eluted through the paper channel with TE buffer and the paper channel was excised into three pieces for the further polymerase chain reaction (PCR) assay. The extraction efficiency was determined by measuring fluorescence reflectance with either a benchtop optical detection system (consisting of an LED light source, a pair of optical fibers, and a miniature spectrophotometer, all built on micro-positioning stages) or a smartphone-based fluorescent microscope (in-house fabricated). The limit of detection of Salmonella Typhimurium in 10% poultry packaging liquid with cellulose paper was 103 CFU mL−1, while that extracted with nitrocellulose paper was 104 CFU mL−1 (as determined by both PCR and fluorescence reflectance). Cellulose channels proved more appropriate for measuring low and very high concentrations of pathogen DNA, while nitrocellulose proved better for analysing the mid-range concentrations. We observed that DNA migrated through nitrocellulose at a faster rate and further than through cellulose due to charge–charge repulsion between nitrocellulose and DNA (both negatively charged), thus contributing to consistent and efficient extraction. We tested the efficiency of Salmonella extraction from 10% poultry packaging liquid, 10% whole blood, and 10% fecal samples, and obtained comparable extraction efficiency, as confirmed by smartphone-based direct fluorescent detection. This protocol is suitable for the direct detection of total bacteria count in a dirty sample (when specificity is not necessary) as well as determining extraction efficiency. This protocol is compatible with PCR, to provide specific information about the type of pathogen present in sample.

 Please wait while we load your content...
Please wait while we load your content...