Programmed assembly of polymer–DNA conjugate nanoparticles with optical readout and sequence-specific activation of biorecognition†
Abstract
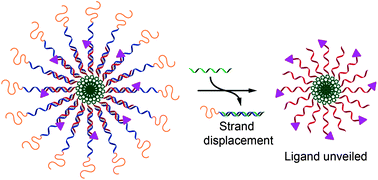
Soft micellar nanoparticles can be prepared from DNA conjugates designed to assemble via base pairing such that strands containing a polymer corona and a cholesterol tail generate controlled supramolecular architecture. Functionalization of one DNA conjugate strand with a biorecognition ligand results in shielding of the ligand when in the micelle, while encoding of the DNA sequences with overhangs allows supramolecular unpacking by addition of a complementary strand and sequence-specific unshielding of the ligand. The molecular assembly/disassembly and ‘on–off’ switch of the recognition signal is visualized by FRET pair signalling, PAGE and a facile turbidimetric binding assay, allowing direct and amplified readout of nucleic acid sequence recognition.
- This article is part of the themed collection: Functional Nanoparticles for Biomedical Applications

 Please wait while we load your content...
Please wait while we load your content...